Maize Genetics Cooperation Newsletter vol 81 2007
KEW, United Kingdom
Royal Botanic Gardens, Kew
SAINT PAUL, MINNESOTA
University of Minnesota and USDA-ARS
Adding B-chromosomes of Zea mays L. to the genome of Avena sativa L.
--Kynast RG; Galatowitsch, MW; Huettl, PA; Phillips, RL;
Rines, HW
B-chromosomes (Bs) are supernumerary dispensable chromosomes described in hundreds of animal and plant species, including maize (Zea mays L.). However, Bs have not been reported to exist in hexaploid oat (Avena sativa L.).
In
order to transfer maize Bs sexually from maize to oat genomes, we chose the
maize cultivar Black Mexican Sweet (a well known sweet corn line hosting Bs in
different numbers) as the B donor (male parent) and the oat cultivars Starter,
Sun II and Paul as potential B recipients (female parent) for inter-species
cross-hybridizations. Since
all of these direct crossings of Black Mexican Sweet to each of the oat
cultivars failed to produce vigorous F1 offspring, we used in a further
experimental series as the male parent a backcross line of the maize inbred B73
harboring Bs from Black Mexican Sweet.
The B73B derivative is the 5th backcross generation of the F1
(B73 �
Black Mexican Sweet) hybrid to B73.
BC5 seeds with hexasomic B addition (BC5-B73B, 2n = 2x+6B
= 26) were generously provided by J. A. Birchler, University of Missouri
Columbia. This genotype based on
B73 germplasm seemed more promising because, recently, Kynast et al., PNAS
101:9921-9926, 2004) had reported the successful crossing of B73 without Bs to
different oat genotypes.
All parental plants were cultivated in growth chambers to deliver favorable environmental conditions for germination and plant growth, and to synchronize the peak of pistil receptiveness in oat plants with the peak of pollen grain release in maize plants. For inter-species crossing, the stigmas of emasculated oat florets were hand-pollinated with freshly shed maize pollen by using a fine camelhair brush. The panicles with pollinated florets were isolated in glassine bags, then 24-48 hours after pollination sprayed with a mixture of 50 ppm 2,4-D and 50 ppm GA3 and again bagged during further cultivation in the growth chambers.
From 2341 ovaries, 115 immature F1 (oat � maize) embryos (Table 1) were in vitro rescued 14-15 days after pollination. The F1 embryos were cultivated on modified one-half strength MS-medium. A total of 31 F1 embryos germinated and developed into vigorous plantlets large enough for molecular and cytogenetic analyses. Plantlets were further grown in growth chambers with optimal growing conditions to produce F2 seeds for testing (1) fertility of (aneu)haploid oat plants with added maize Bs and (2) transmission of maize Bs to the offspring in an oat background.
Table
1. Plant material for crossing
three different oat cultivars (2n = 6x = 42) by the maize B73B (2n =
2x+6B = 26) and results of maize B-positive offspring production.
|
Oat
cultivars |
Starter |
Sun II |
Paul |
Total |
|
Oat
panicles |
40 |
53 |
3 |
96 |
|
Oat
florets, emasculated and hand-pollinated |
1177 |
1094 |
70 |
2341 |
|
F1
proembryos, rescued 14-15 dap* |
62 |
52 |
1 |
115 |
|
F1
embryos, geminated** |
14 |
16 |
1 |
31 |
|
Maize
(A and/or B)-positive juvenile F1 plantlets (shoot- and root-tested) |
7 |
6 |
1 |
14 |
|
Maize
B-positive adult F1 plants*** (tiller-tested) |
2 |
0 |
0 |
2 |
|
Maize
B-positive F2 offspring / Total F2 offspring (shoot- and root-tested) |
20 / 30 |
0 / 0 |
0 / 0 |
20 / 30 |
*Days
after pollination; **Embryos that formed shoot and root with enough tissue for
molecular and cytogenetic analyses; ***Plants represent tillers that are clone
parts of two clones after extensive tiller cloning of both F1 plants allowing
for more F2 seed production
Two F1 plantlets (5811-1 and 5845-1) were found to have retained maize chromosomes in shoot tissues based on results from a PCR assay for Grande1, a dispersed LTR-type retrotransposon which is abundant on all A chromosomes (As) and Bs of maize, but absent from all chromosomes of the oat genotypes used in our crossing program. PCR assays involving two B-specific markers (primer pair p-2ndB1 + p-2ndb4 and primer pair p-brpt2 + p-taralb1; generously provided by J. A. Birchler, University of Missouri Columbia) and a selected set of A-specific markers for maize [chromosome arm-specific SSR markers selected from the �Maize Genetics and Genomics Database� (http://www.maizegdb.org/)] showed that in both genotypes the Grande1-positive PCR products represented the presence of maize Bs and not maize As (Fig. 1).
Cytological analyses on very young, juvenile plantlets revealed that in the F1 plant 5811-1, all ten maize As had been eliminated and a complete set of 21 oat chromosomes plus three maize Bs (2n = 3x+3B = 24) were retained in its primary root meristem. In the primary root meristem of the F1 plant 5845-1, all ten maize As had been eliminated with a complete set of 21 oat chromosomes and a single maize B retained (2n = 3x+1B = 22).
Both
F1 plants were kept under short-day conditions to allow
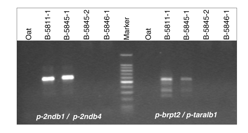
Figure
1. PCR products of
B-chromosome-specific markers after electrophoresis in 1.5% agarose; both
markers demonstrate the presence of B-chromatin in the two F1 plants 5811-1 and
5845-1 and the absence of B-chromatin in two examples of B-negative F1 plants
(5845-2 and 5846-1).
plants to tiller extensively for continuing tiller cloning. Tiller cloning provides an extended source of leaves for the extraction of genomic DNA and RNA, and for further seed production. Both genotypes are descended from a (Starter � B73B) cross. Hence they represent B additions in a Starter background. The phenotypes of both mature F1 plants did not differ from those of haploids of Starter without Bs at any point in time during their growth period.
After shifting individual tiller clones into long-day growing conditions, self-pollination produced F2 seed of both genotypes (Table 1). This fertility could be attributed to the frequent formation of numerically unreduced female and male gametes. High fertility had already been observed in oat haploids of Starter, Sun II and Paul without and with individual maize As of B73 (Rines et al., In: Jain, Sopory, Veilleux (eds.) Kluwer Acad. Publishers, Dordrecht, The Netherlands, In vitro haploid production in higher plants 4, 205-221, 1997; Kynast et al., 2004).
Cytological and molecular analyses of 20 F2 offspring plants showed that the F1 plant 5811-1 carrying 3 Bs produced three F2 plants with 1 B, six F2 plants with 2 Bs, one F2 plant with 3 Bs, one F2 plant with 4 Bs, and nine F2 plants with highly chimeric root meristems showing cells with 1-5 Bs in different frequencies. All chromosome counts were based on ten cells of root meristems of each F2 offspring. The presence of Bs in root meristem cells was visualized by GISH at high (85%) stringency using Alexa Fluor 488-labeled genomic DNA of maize as the probe without oat competitor DNA (Figure 2).

Figure
2. Root prometaphase cell from the
F2 plant K1188; the tetrasomic addition of maize B-chromosomes to Starter oat
is demonstrated by green fluorescence after GISH with Fluor 488-labeled genomic
DNA of maize well contrasted against the red-brown counterstained oat
chromosomes. (For color see online.)
For the F1 plant 5845-1 carrying one B, none of the 10 F2 offspring tested by cytological and molecular means had Bs, indicating a transmission failure. However, since more F2 seeds were produced, we will analyze a larger offspring population of F1 plant 5845-1 to evaluate the transmission data.
Taking all the data together, our results showed that Bs from maize Black Mexican Sweet can be sexually transferred to the genome of Starter oat, and that haploid oat plants hosting one or three maize Bs are fertile and can set seed after self-pollination. In addition, our results of 30 tested F2 offspring from two maize B-positive F1 plants showed that maize Bs can be transmitted to offspring even when in the presence of only oat chromosomes.
Please Note: Notes submitted to the Maize Genetics Cooperation
Newsletter may be cited only with consent of authors.