The nucleotide sequences of ORF3 in a VIR42 MV hybrid and from the genotype reported earlier are almost identical. The only difference was found in position 963 of the sequence (guanine instead of cytosine). This minor alteration in nucleotide sequence leads to amino acid substitution (histidine � aspartic acid) in the polypeptide product of ORF3 (see Fig. 2).
The results of sequencing of an ORF1 fragment in the S2 plasmid in a VIR42 MV hybrid are shown in Figure 3.
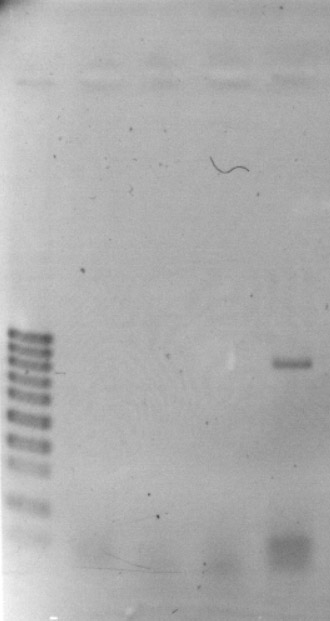
Two nucleotide changes in the ORF1 sequence in the S2 plasmid of a VIR42 MV hybrid were revealed (T � A and G � A). Both S1 and S2 plasmids are in the integrated state in this hybrid, but the 2.3 kb linear plasmid behaves as an autonomous replicon. The ORF1 transcript in the S3 plasmid was detected by RT-PCR (Fig. 4). At the moment we are sequencing this RT-PCR product. We have been unable to detect the ORFs product in S1 and S2 plasmids in this hybrid.
The data presented demonstrate that ORFs in linear plasmids of maize mitochondria in different genotypes have a rather conservative nature.
tctagaa actcttttat taaaaagacg agacacggat gtcgacaaaa 900
******* ********** ********** ********** **********
ctcacgtgcc ctacgcaggg gggtatatga tggttgatat ggaaaagcgg gttaacgcgg 960
********** ********** ********** ********** ********** **********
accatattac gacgttctat gcacatgact actcaaaagt gtgccaggat tttcacgata 1020
**g******* ********** ********** ********** ********** **********
tgagtgaaaa gatgctcaca gagatgatta atagaatagt aaaggatgtt caaagaagag 1080
********** ********** ********** ********** ********** **********
gaagttcaat ggttgtatac tttcacaatt tatctcagtt cgacggtatt atgatactct 1140
********** ********** ********** ********** ********** **********
cttttttaac taaaagttat aaaaactgtc atatagagcc catcatgaga aacgactgta 1200
********** ********** ********** ********** ********** **********
tttattccat aaaactgtat aaggtatcca aaaatgggga taaaaggctt gttttaacgt 1260
********** ********** ********** ********** ********** **********
tcatggattc gtacctcctg ttgaaagtaa agctcgctga tctagccgat agtttttgcc 1320
********** *********
Figure 1. The nucleotide sequence of ORF3 in S1 plasmid of a VIR42 MV
hybrid. The strand shown corresponds to the EMBL Database sequence accession
X02451. Primers for PCR amplification of the ORF3 region used were the
following: 5�-primer, 5�-AGAAATGAAAAACAAAACTC-3�; 3�-primer, 5�-CAAAAACTATCGGCTAGATCAG-3�.
Purified double-stranded amplification product was sequenced using the
dideoxy chain termination method (Sanger et al., 1977) as described in
Promega�s Technical Protocol. The nucleotide sequence was determined on
both strands of DNA in triplicate. The products of sequencing reactions
were resolved by 5 and 8% polyacrylamide-8 mol/L urea gel electrophoresis,
and autoradiographed for 24 hours at room temperature.
VADLETLLLKRRDTDVDKTHVPYAGGYMMVDMEKRVNADHITTFYAHDYSKVCQDFHDM
***************************************D*******************
SEKMLTEMINRIVKDVQRRGSSMVVYFHNLSQFDGIMILSFLTKSYKNCHIEPIMRNDC
***********************************************************
IYSIKLYKVSKNGDKRLVLTFMDSYLLLKVKLADLADSFCPELGGKGSFDHQNVTVDKL
***********************************************************
Figure 2. Deduced amino acid sequence of ORF3 product in a VIR42 MV
hybrid.
agtct
*****
1741 atgattttat acaaaaacat tggagtgtat tagtgtccgt ggggcttctc aggccgaaga
********** ********** ********** ********** ********** **********
1801 atctagcttt atttaaacga aaggaggcgc tcaggctact atctagcctt ttgtttaaac
********** ********** ********** ********** ********** **********
1861 acgaggagct ttcaacgatt tatcgatata gtgagttgaa atctgtattg ttaaaaaata
********** ********** ********** ********** ********** **********
1921 tacacgcgtc aaccttcgaa ctatatacta tgaaaatagc tgaggcttat ctagattata
********** ********** ********** **********
********** **********
1981 aaatctattt tccaatcttt ctggacttca gggggcgaaa ttaccgccat ggacccttcc
********** ********** ********** ********** ********** **********
2041 atttccacga acgtgattta gtgagatcac tcatcatatt tgatgaaagt gatgactcag
********** ********** ********** ********** ********** **********
2101 cagcacatac tataaatagt gatgttgggg atagaatcct ccataatttc cttatatcag
********** ********** ****a*a*** ********** ********** **********
2161 cgg
***
Figure 3. The nucleotide sequence of ORF1 in the S2 plasmid of a VIR42 MV hybrid. The strand shown corresponds to the sequence from EMBL Database (EMBL J01426).

M 1 2
Figure 4. Electrophoretic analysis of the RT-PCR product of ORF1 in
2.3 kb plasmid-like DNA in a VIR42 MV hybrid. 1, control; 2, RT-PCR product.
M, molecular weight markers.
Return to the MNL 77 On-Line Index
Return to the Maize Newsletter Index
Return to the MaizeGDB Homepage