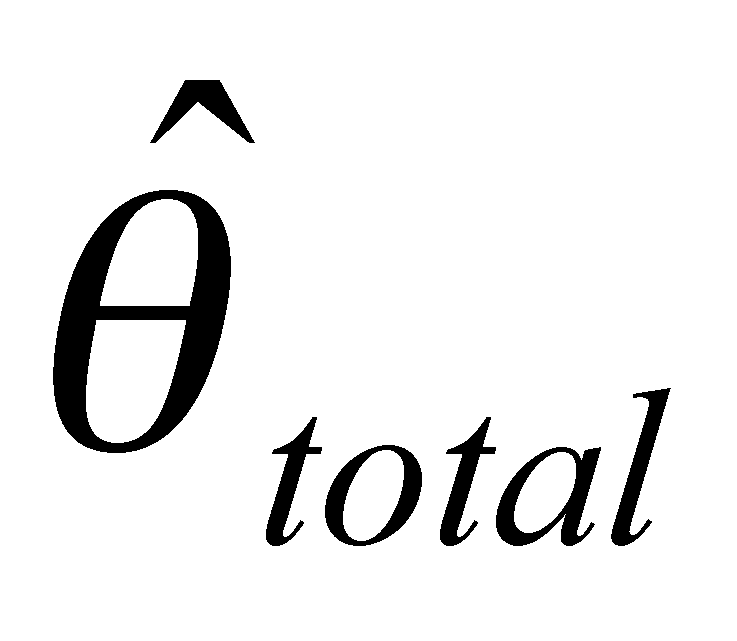
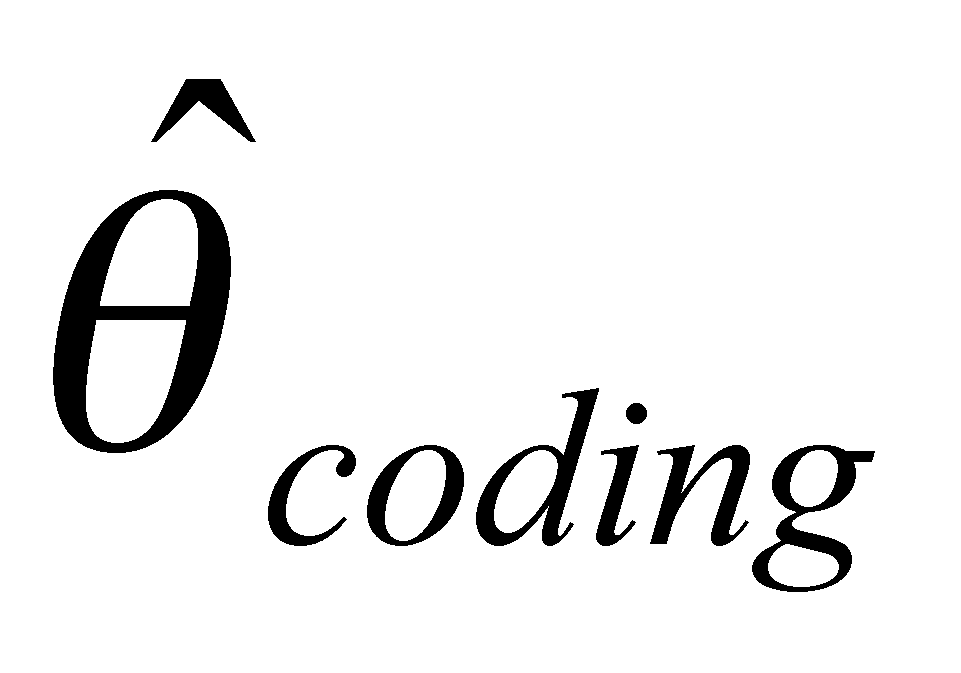
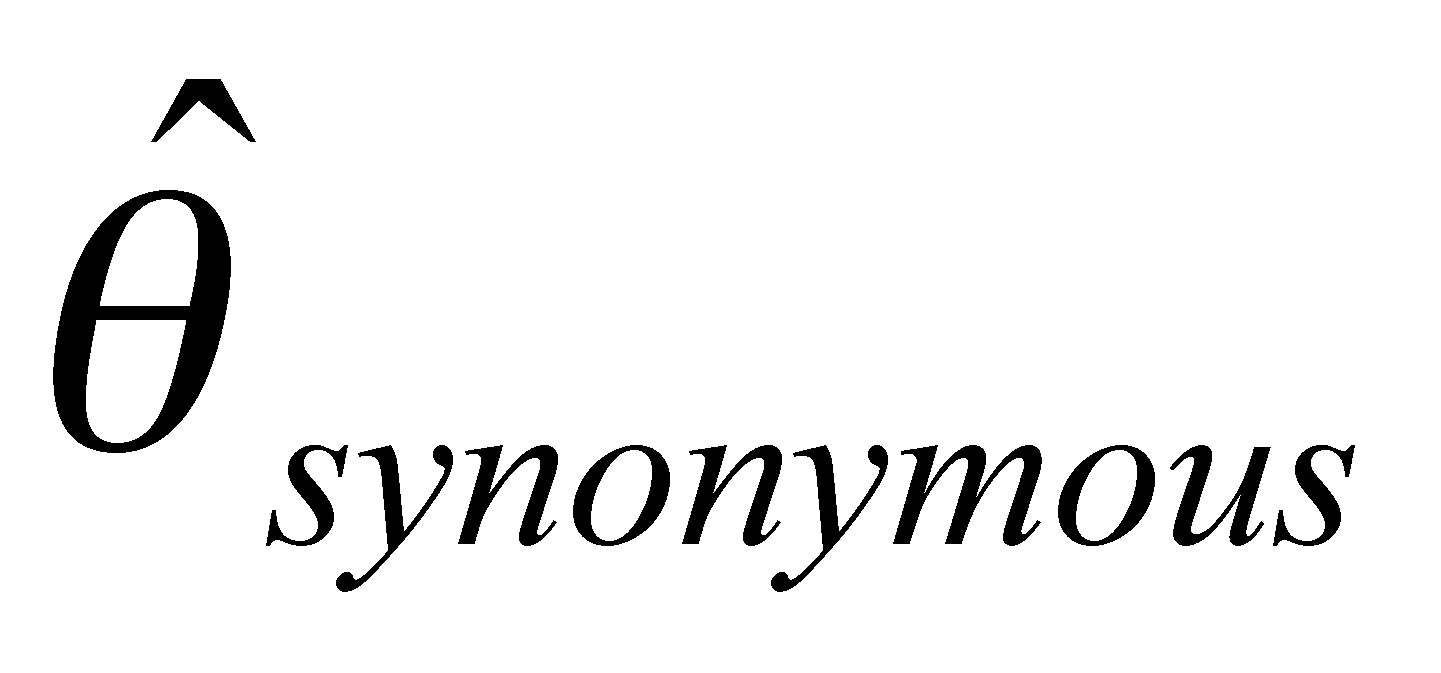
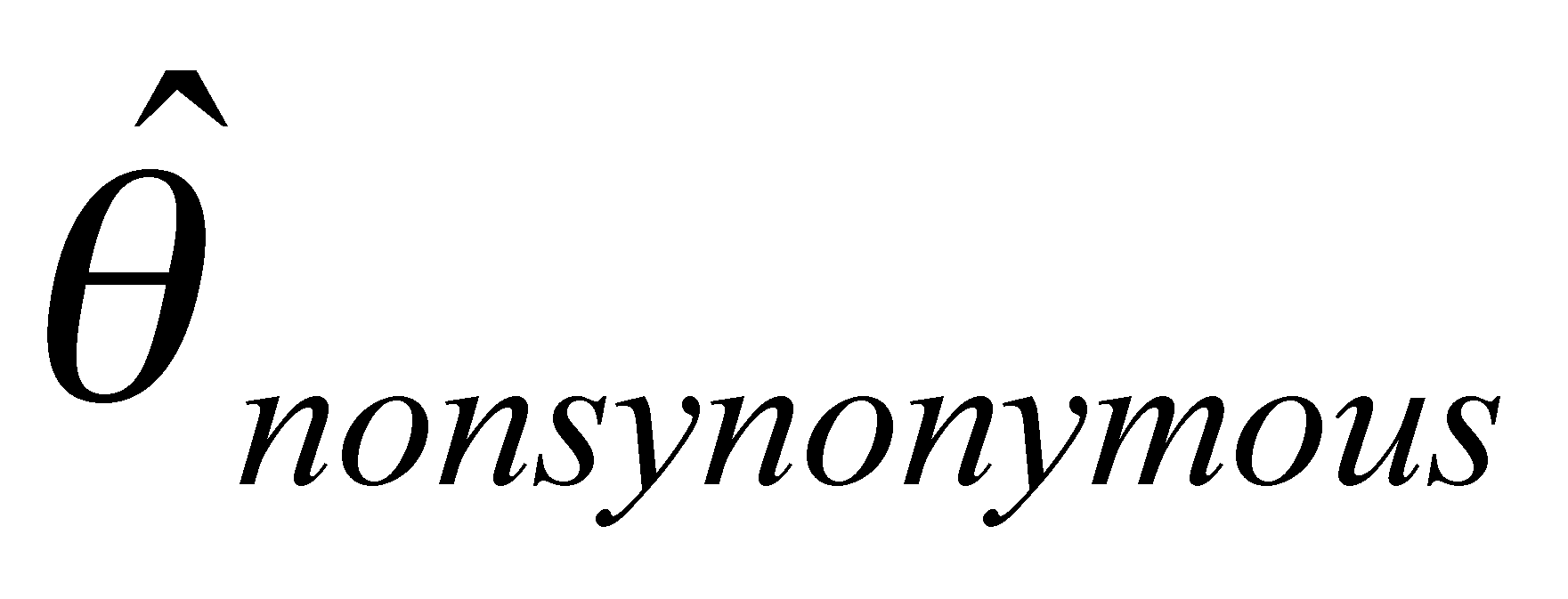

Our project is composed of three data-generating laboratory groups plus a statistical genetics support group. Group 1, led by John Doebley and Steve Kresovich, is examining microsatellite diversity in maize. Group 1 employs microsatellites in part because they are relatively inexpensive, allowing a large and comprehensive sample of the maize germplasm pool and the entire maize genome to be assayed for genetic diversity. Group 2, headed by Brandon Gaut, is examining DNA sequence diversity in maize with a focus on a limited set of accessions and regions of the genome. Group 3, headed by Ed Buckler, seeks to identify polymorphisms within candidate genes that influence agronomically important traits. To do this, the Buckler group draws on the knowledge of diversity in maize generated by the first two groups since one cannot safely make inferences about the associations between genotypes and phenotypes without incorporating a knowledge of the distribution of diversity into the analysis. The project includes a fourth group headed by Spencer Muse and Bruce Weir. This group provides statistical support for the three data-generating groups and is designing and implementing a database for genetic diversity in maize. Finally, Major Goodman is assisting the data generating groups by selecting and providing appropriate germplasm samples for their analyses and working with Ed Buckler's group on the phenotypic analyses.
Below we summarize the goals and activities of the four groups within our project to date.
Group 1: Microsatellite Diversity In Maize. This portion of the project is investigating the evolutionary dynamics of maize microsatellites and exploiting microsatellites to characterize genetic diversity in maize. We are addressing the following questions.
(1) Do maize microsatellites follow a stepwise mutation model? Microsatellites are simple sequence repeats (SSRs) that in theory evolve in a stepwise manner by changes in the number of repeat units. For example, a trinucleotide (CCG) repeat SSRs should have alleles that differ from each other by multiples of 3 bp. Our results show that maize microsatellites do not follow the stepwise mutation model. Rather, we found that there is a continuous distribution of allele sizes for most maize microsatellites. To answer why this is the case, we sequenced a selection of maize and teosinte alleles at several microsatellites and learned that most of the variation is due to indels in the regions flanking the microsatellite repeat (Matsuoka et al., Theor. Appl. Genet., in press, 2002). Thus, the reason for this violation of the stepwise mutation process is that most variation at maize microsatellite loci is due to indels in the flanking regions and not changes in the number of repeats.
(2) At what rate do microsatellites evolve? We have found that mutation rates for maize microsatellites are similar to those of humans and yeast but higher than those of Drosophila. We determined the mutation rate for SSRs by assaying 100 SSRs for 86 RI lines after 11 generations of selfing (about 80,000 allele-generations). The mutation rate for dinucleotide SSRs was 4.4 x 10-4 for repeat length changes. Among these lines, we observed no indels in the flanking regions, indicating the different dynamics for repeat length changes and indel mutations during short-term evolution. Also, SSRs with trinucleotide repeats or higher produced no mutations in our mutation accumulation experiment showing that these classes of SSRs evolve at a much slower rate (Vigouroux et al., Mol. Biol. Evol., in review).
(3) How is diversity in the maize germplasm pool distributed? Native Americans distributed and adapted maize from Chile to Canada. We are using microsatellites to understand how diversity is structured among native landraces, US breeding lines and maize�s wild relative (teosinte). We have screened over 1200 landraces and 200 wild maize (teosinte). An initial analysis of a comprehensive sample of maize landraces revealed that maize genetic diversity is structured largely along eco-geographic lines (Matsuoka et al., Proc. Natl. Acad. Sci. USA, in review) and that maize was domesticated from teosinte only once. This analysis also indicated that the maize exotic germplasm pool can be divided into three supragroups: North American Maize (maize of the Indian tribes of the US and Canada), tropical maize (most maize of Mexico, Central and lowland South America), and Andean maize (maize of the Andes Mountains growing above 2000 meter elevation).
(4) How is microsatellite diversity distributed across the maize genome? We seek to determine how factors such as the domestication bottleneck and proximity to genes under selection during domestication have sculptured SSR genetic diversity in the maize genome. We have found that diversity is not highly patterned across the genome. For example, SSR diversity in regions of the genome that harbor domestication QTL is not substantially lower than in other regions. Nevertheless, our analyses have identified several dozen SSRs in genic regions that appear to have been targets of selection during domestication. Over all SSR types, maize possesses only 81% of the SSR allelic diversity seen in teosinte, demonstrating that SSRs have yet to fully recover from the domestication bottleneck.
Group 2: Processes that Shape Genetic Diversity in the Maize Genome. Patterns of genetic diversity can provide insight into evolutionary processes that shape genomes, like mutation, recombination, and natural selection. Brandon Gaut's group is studying DNA sequence diversity in maize and its wild relatives to elucidate processes that have shaped the evolution of the maize genome. Thus far, they have characterized single nucleotide polymorphisms (SNPs) in 21 loci along chromosome 1, based on a sample of 25 individuals representing 16 exotic landraces and 9 U.S. inbreds (Tenaillon et al., Proc. Natl. Acad. Sci. USA 98:9161-9166, 2001). These data indicate:
A. Among the organisms that have been well-characterized to date at the DNA sequence level, maize contains the most SNP diversity (Table 1). On average, two randomly chosen maize sequences will vary at 1 out of 104 base pairs.
B. U.S. inbreds retain only 77% of the SNP diversity of exotic landraces, indicating a loss in diversity due to selective breeding. Surprisingly, the same inbred sample retains >95% of the genetic diversity of landraces at microsatellites (SSRs) within the 21 chromosome 1 loci. Differences between SNP and SSRs may be due to rapid SSR mutation rates that mask historical diversity-reducing events.
C. There is no obvious pattern of SNP diversity along the genetic map of chromosome 1. For example, in Drosophila, it has been shown that loci near the centromere are less variable than those distant from the centromere. This relationship does not appear to occur for SNPs in maize.
Currently, the Gaut laboratory is investigating the relationship between diversity and recombination. Diversity and recombination is correlated in several organisms, but the relationship is not straightforward in maize. We believe the relationship has been obscured by the genetic effects of domestication. This theory will be addressed in our studies of wild relatives.
Group 3: Relating Nucleotide Diversity to Phenotypic Diversity. Ed Buckler's group is pioneering the application of association analyses to plants. Association analyses seek to measure whether polymorphisms in a candidate gene are correlated (or associated) with phenotypic variation for traits that the gene is known to influence. Association approaches are fast and provide high resolution, but they had not been applied to plants due to problems with population structure, linkage disequilibrium (LD), and sequencing costs. We are trying to overcome some of these obstacles by incorporating factors such as population structure and LD into the statistical models used.
An initial issue addressed by the Buckler group was to document the extent of linkage disequillibria in maize. Understanding LD is the key to determining the possible genetic resolution of candidate gene association approaches in maize. Linkage disequilibrium is the correlation between polymorphisms across a sample. By sequencing 18 candidate genes from 102 diverse maize inbred lines, it was found that LD generally decays very rapidly in most loci. LD generally decays within 1500bp (Remington et al., Proc. Natl. Acad. Sci. USA 98:11479-11484, 2001).
Another important issue in association analyses is population structure. If genetic diversity is highly structured among subpopulations, it can lead to false associations between genes and phenotypes. The Buckler group was able to control statistically for this bias by using SSR genotypes provided by our microsatellite group. Essentially, they constructed a model that first factored out associations between genes and phenotypes due to population structure as measured by SSRs before testing for associations between genes and phenotypes.
The Buckler group first investigated candidate genes for flowering time (Thornsberry et al., Nature Genetics 28:286-289, 2001). By sequencing the Dwarf8 gene from a set of 92 maize lines, they could test whether individual nucleotide polymorphisms are associated with a large effect on flowering time. This research also involved the development of statistics to handle population structure, which had been a major confounding factor in many animal and human studies. They found three important polymorphisms that were significantly associated with flowering time. Of particular interest is the 6bp deletion flanking the SH2 domain. The development of novel statistical approaches was key to our analysis, and is why this research will attract attention in many other organisms.
Group 4: Statistical Genetics and Database Development. The data being generated by the first three groups poses some novel analytical problems and the need for an innovative database that will allow the maize genetics community to make full use of the data and results. Spencer Muse and Bruce Weir are working on these issues.
To assist in data analysis, the Muse group has developed data analysis tools, such as PowerSSR, which is designed for evolutionary analysis using SSR data (see www.stat.ncsu.edu/ ~panzea/software/software.html). PowerSSR has a variety of attractive features and capabilities. It features an Internet Explorer-like user interface. It performs the following analyses: basic data description, allele frequency estimation, 17 genetic distance measures, phylogenetic analysis, population structure analysis, and linkage disequillibrium analysis. It includes a hierarchy editor that allow up to 4 hierarchical levels for population structure analysis. PowerSSR reads and exports multiple data formats including Excel, Nexus and Arlequin.
The Muse group is also working to develop a genetic diversity database
for maize called Panzea (www.stat.ncsu.edu/~panzea/).
This database will allow project members and the community in general to
create datasets that combine the disparate data types generated by our
project, including DNA sequences, SSRs and phenotypes. Web-based data submission
tools allow project members to submit data into our databases. The basic
scheme and design of the database have been implemented and the current
focus is on developing a variety of graphical search tools targeting "major"
categories of work, including genetic diversity, genotype-phenotype association
and geography. One tool under development is a genetic map based portal
into Panzea. Here, the user could select a chromosome (or set of chromosomal
bins) and then view a genetic map of that chromosome with the loci along
its length and measures of genetic diversity at each locus. The user could
then click on an individual locus to view or down load the data for further
analysis. Other tools would allow the user to enter the database by first
viewing a geographic map of maize varieties and then view or download diversity
data based on geographic criteria. Finally, a phylogenetic tool would show
the user a phylogeny for maize varieties and allow the user to view or
analyze diversity or phenotypes for the selected varieties.
Table 1. A comparison of sequence diversity among three species. Human
estimates are based on two different studies, corresponding to the two
columns, that varied in the genes sampled. Values for (a measure of diversity)
indicate that maize is 11 times more variable than humans and 1.4 times
more variable than D. melanogaster.
|
|
|
|
||
| No. of loci |
|
|
|
|
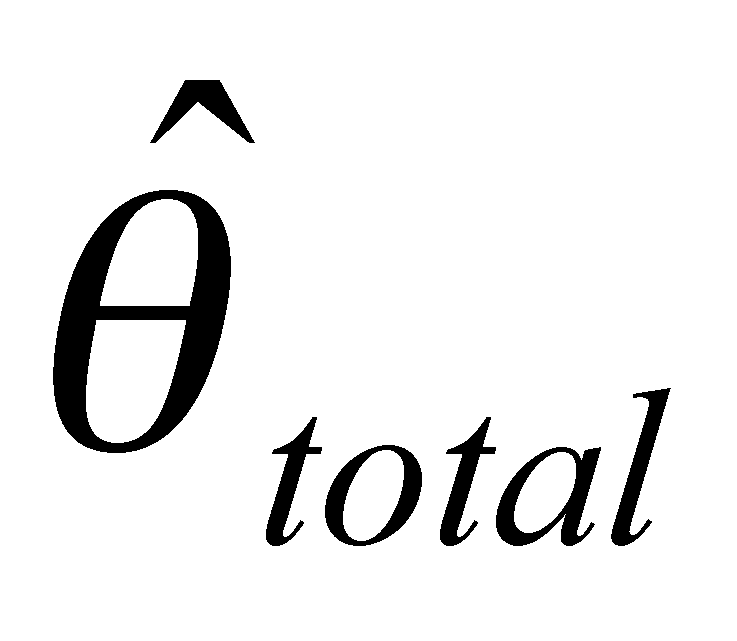 |
|
|
|
|
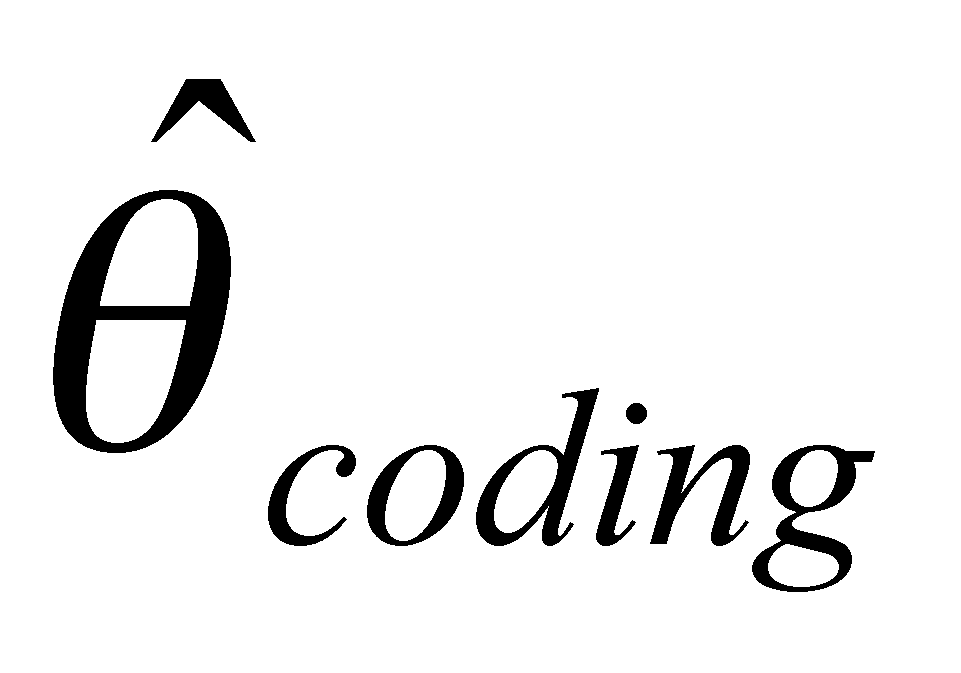 |
|
|
|
|
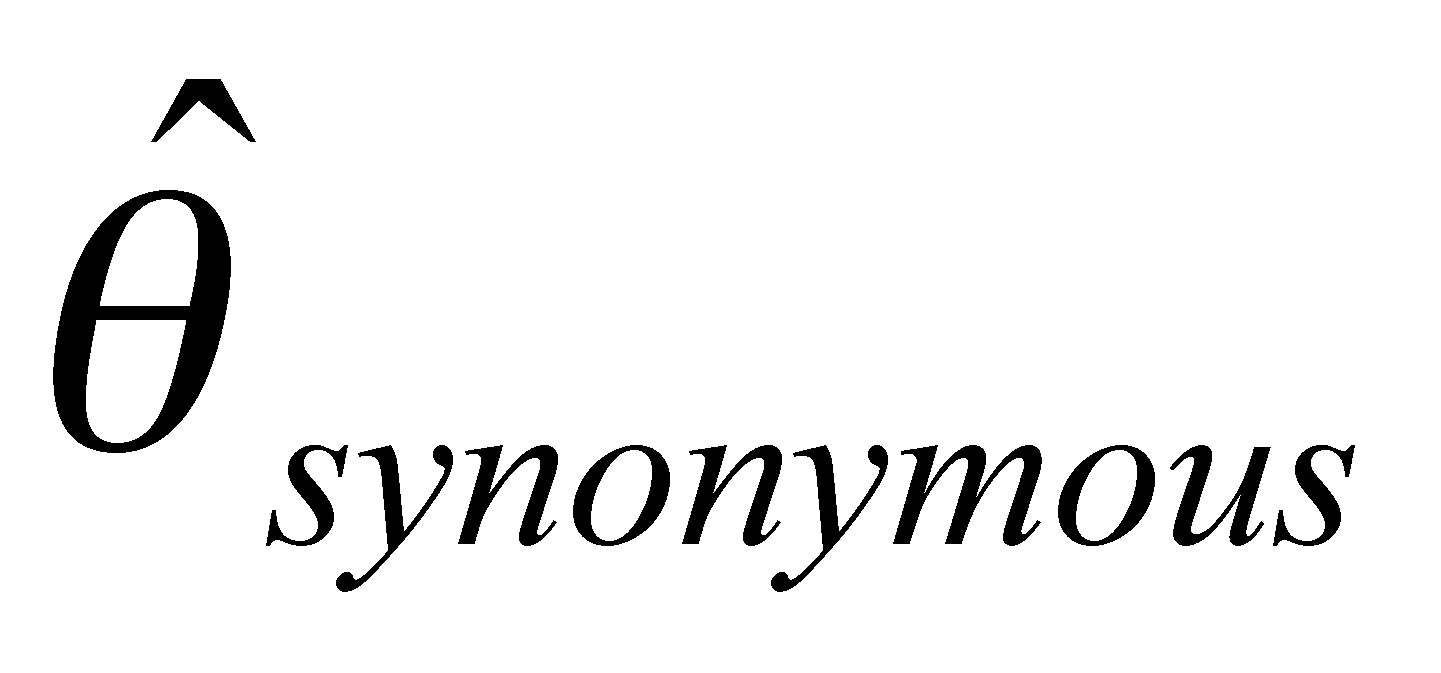 |
|
|
|
|
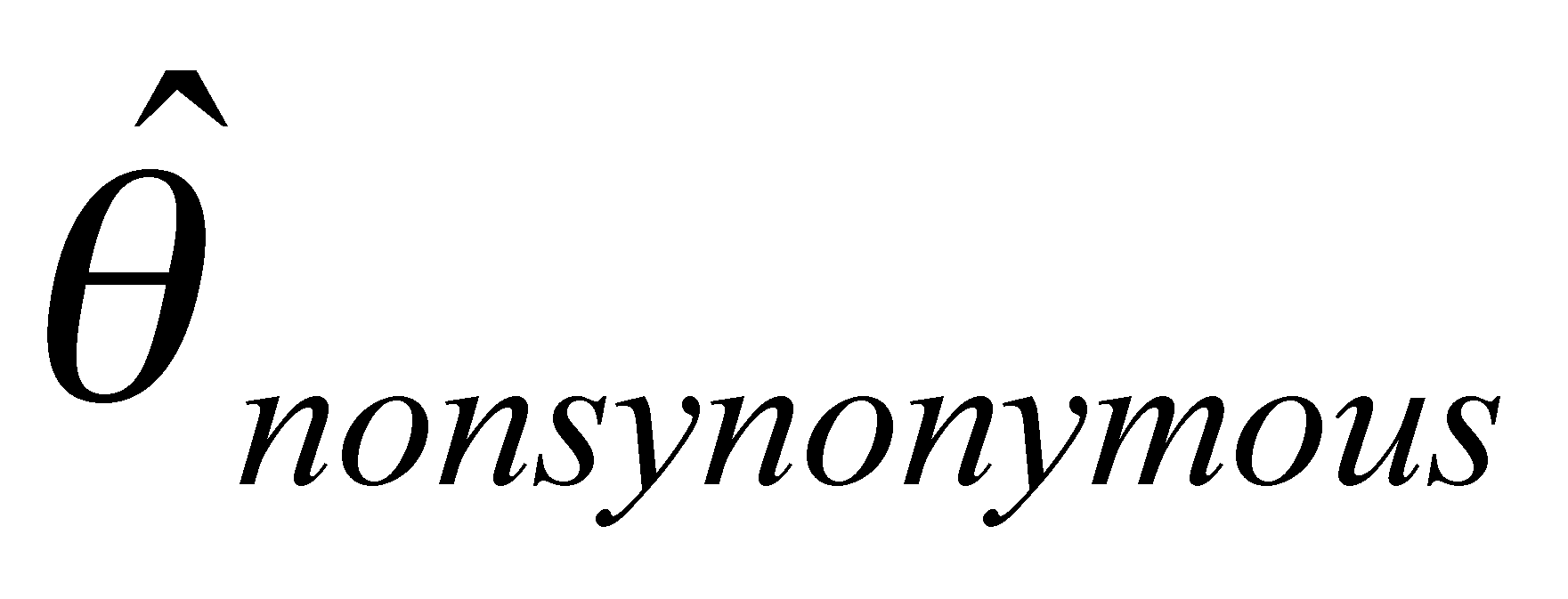 |
|
|
|
|
 |
|
|
|
|
Return to the MNL 76 On-Line Index
Return to the Maize Newsletter Index
Return to the MaizeGDB Homepage