Maize Genetics Cooperation Newsletter
vol 87 2013
PISCATAWAY, NEW JERSEY1
Waksman Institute, Rutgers University
MONTCLAIR, NEW JERSEY2
Montclair State University
A
sequence-indexed single gene knockout resource for maize
Li, Y1; Huang,
J1; He, L1; Wang, Q1; Xiong,
W2; Segal, G1; Du, C2;
Dooner, HK1
The purpose of this note is to apprise the maize genetics community of
progress in our NSF-funded project to develop a single-gene-knockout reverse
genetics resource based on the transposon Ds. We have three objectives: (1) To construct a set of 120 roughly
equidistant transgenic Ds launching platforms that will allow simple
visual selection of element transposition from any region of the genome and,
thus, enable researchers to generate regional gene knock-out collections, (2)
To isolate several thousand Ds insertion sites from
model platforms and sequence-index them using a combination of next-generation sequencing
(NGS) technology and computational tools that should make the method generalizable
to any collection of insertions produced in a common background; and (3) To
develop a web-searchable database of insertion site sequences cross-referenced
to stocks available from the Maize Genetics Stock Center.
The
transposons Ac and Ds tend to insert
in or close to genes and are, therefore, excellent gene-searching
engines in the highly repetitive maize genome (Cowperthwaite
et al., Plant Cell 14:713-726, 2002; Vollbrecht et al., Plant Cell 22:1667-1685, 2010). Both elements show a distinct preference to transpose to linked sites: about
one-third of all transpositions are within 7 cM on
either side of the donor site (Greenblatt, Genetics 108:471-485, 1984; Dooner and Belachew,
Genetics, 122:447-457, 1989; Cowperthwaite et al., Plant Cell
14:713-726, 2002). Therefore, Ac/Ds elements are excellent gene tagging tools for localized
transposon mutagenesis and complement Mutator elements, which show a more random pattern of
insertion across the genome.
In our
project, we transform the c1 HiII hybrid with Agrobacterium using a standard
binary vector system (Frame et al., Plant Physiology 129:13-22, 2002). Our
engineered construct is based on the c1-m2 mutable allele originally
described by McClintock (Carnegie Inst Wash Yrbk 47, 155-169, 1948), in which a Ds element is inserted
in the third exon of the c1 gene
(Cone et al., Proc Natl Acad Sci USA 83:
9631-9635, 1986). In the presence of Ac, c1-m2 shows a spotted
aleurone phenotype. We
have modified the Ds element to
include a GFP marker expressed behind
a 22-kD zein promoter that allows us to trace the
movement of the element in the genome (Figure 1).
As outlined in
Figure 2, when test crossed with a colorless Ac donor line, most transgenotes showed
kernel spotting. The c-m spotted
phenotype resembles that of the native c1-m2 allele and segregates 1:1,
suggesting transgene integration at a single locus. Furthermore, all spotted kernels are green
fluorescent, confirming that the spots are due to transpositions of Ds:GFP
in response to Ac. Southern-blot and progeny analyses
confirmed the integration, expression, and inheritance of the transgenes in the
T1 and T2 generations. Sequences adjacent
to the T-DNA launching platforms were isolated by inverse PCR, sequenced, and
mapped to the B73 reference genome. So far, we have generated 160 active
transgenic lines and mapped 82 platforms to the maize genome. Their location and those of any new
mapped platforms can be found in our project website, http://www.acdsinsertions.org. These lines are being deposited
in the Maize Stock Center and will serve as starting materials for the
generation of gene knockouts by community researchers.
We have measured
the reversion frequency of each platform by crossing to a colorless c1 tester and selecting for C� revertants. The
average reversion frequency was 2.97 x 10-2 on the male side and
1.64 x 10-2 on the female side, a bit higher than that of the native
c1-m2 allele (2.0 x
10-2 as male, and 0.7 x 10-2
as female). The higher reversion frequency makes
these lines more efficient for generating mutations.
To date, more
than 12,000 purple C� revertants have been selected
from nine platforms and those from three platforms have been further characterized
genetically. The GFP marker in the c1-m2
(Ds) allele serves to track Ds* reinsertion
after excision from the c1 gene. The C� revertant
kernels carrying a trDs* are readily distinguished from those
without Ds under blue light illumination
in a fluorescence dissecting microscope. The average reinsertion frequency is
close to 50%, similar to what has been observed in nontransgenic
systems.
We use the c1-m2 transgenic lines as pollen donors
because about 93% of C� selections from the male side
are concordant (C� endosperm and embryo), whereas only 1/3 of C� selections
from the female side are concordant, i.e., heritable. The genetic
distance between the trDs*
and the C� allele in the T-DNA can be
readily obtained by scoring the fraction of green fluorescent colorless kernels
in the testcross progeny. As
expected, the majority (74.1%) of the genetically linked reinsertions are tightly linked (within 7 cM)
to the original T-DNA platform. 80%
of the tightly linked sites fall between 0 and 3.5 cM.
The trDs*
elements are mapped to the reference genome by isolating and sequencing the reinsertion
sites. To do so, we adopted the splinkerette-PCR method (Uren AG, et al., Nature Protocols
4:789-798, 2009) and constructed libraries for next generation sequencing (NGS)
of pooled trDs*
insertion sites. In brief, genomic
DNA of seedling tissue from C� GFP revertants
arranged in 3-D pools is sheared to 2-3 kb and ligated
with a double stranded splinkerette oligonucleotide. The ligation
products are amplified by nested PCR, in which the first reaction is carried
out with a GFP primer and an adaptor
primer that can only anneal to a template synthesized by linear amplification with
the GFP primer. The nested reaction is
carried out with a primer from the end of Ds
and a barcoded adaptor primer. The
amplified insertion junctions are sequenced in-house by a SOLiD
5500xl system.
At present, we have used the above protocol on two sets of 960 C� revertants arranged in 3-D pools of 10 plates x 8 rows x 12
columns. . A new pipeline, InsertionMapper, was
specifically developed
for the project. Using this pipeline,
we have been able to assign trDs* junction sequences
to 1320 individual C� revertants and anchor them to
the reference genome. Among them,
1183 are inserted in single copy DNA, and 809 are in genes. The results of the physical mapping are
generally consistent with those of the genetic mapping. All the above information is provided in
our website, http://www.acdsinsertions.org, and updated
periodically. The
website offers
BLAST search capabilities for researchers to identify the stock(s) of
interest. The stocks will be
available from the Maize Genetics Stock Center, upon
advance APHIS notification of interstate movement.
Acknowledgment. This project is supported
by NSF-PGRP grant DBI 0929350.
Figures
Figure
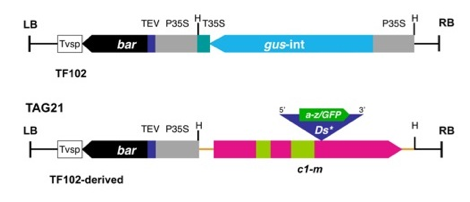
1. T-DNA construct used in Agrobacterium transformation
of Hi-II (c1) embryos. The gus HindIII (H) fragment of pTF102 was replaced with the c1-m Ds*
excision reporter shown in TAG 21 (LB, left border; RB, right border).
Figure
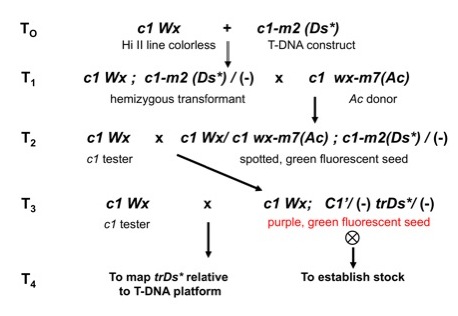
2. Genetic scheme to generate transgenic c1-m lines and isolate C�
revertants carrying a trDs* element
Please Note: Notes submitted to the Maize Genetics Cooperation
Newsletter may be cited only with consent of authors.