Maize
Genetics Cooperation Newsletter vol 87 2013
Buenos Aires, Argentina.
IEGEBA-CONICET and �LACyE, Departamento
de Ecolog�a,
Gen�tica y Evoluci�n, Facultad de Ciencias Exactas
y Naturales, Universidad de Buenos Aires. �
Karyotypical
studies of two maize races from Northeast Argentine (NEA): DAPI-banding and
Fluorescent In Situ Hybridization (FISH)
Realini MF*; L Poggio and GE
Gonz�lez
����������� Knobs� heterochromatic blocks occur in all Zea
species with 2n = 20,
varying in size and number across maize races and their wild relatives (Kato, Mass. Agric. Exp. Stn. Bull.
635: 1-185, 1976; McClintock et al., Colegio de Postgraduados, Chapingo,
M�xico, 1981; Gonz�lez & Poggio, Genome
54: 26–32, 2011; Poggio et al., Cytogenet. Genome Res. 109: 259–267, 2005). Variation in DNA content has been proposed to be principally
due to differences in the amount of heterochromatin, which is mainly located in
distal knobs (Laurie &
Bennett, Herediy 55:307-313, 1985; Poggio et al., Ann.
Bot. 82:115-117, 1998). These structures can be observed as subtelomeric bands by DAPI-banding, and using Fluorescent In
Situ Hybridization (FISH) (Gonz�lez et al.,
Chrom. Res.
14: 629-635, 2006). Knobs have
been described as highly repeated tandem
arrays of 180-bp and �TR-1 (350 bp)
sequences, both repeated in different proportions constituting different knobs (Dennis & Peacock, J. Mol. Evol. 20: 341-350, 1984; Ananiev et al., Proc. Natl. Acad. Sci. 95: 10785-10790, 1998).
In this study we present the karyotypic formulae, asymmetry indexes and the position and
composition of the knobs of two races
of maize from Argentine Northeast (NEA), Tupi
Amarillo (VAV 6563) and Rosado (VAV 6565). DAPI-banding and FISH
techniques were applied. The plant material has been provided by the Vavilov Lab. of Universidad de Buenos Aires (UBA), and
cultivated in the greenhouse of the Facultad de Agronom�a, UBA.
DAPI-banding was performed
according to Summer (Chromosomes banding. Unwin Hyman, London, 1990). DAPI fluorochrome (4�-6-diamidino-2-phenylindole) preferentially
stains AT-rich heterochromatin in plants (Guerra, Genet. Mol.
Biol. 23: 1029-1041,
2000).
The 180-bp and TR-1 knobs sequence of maize was obtained
from GenBank (http://www.ncbi.nlm.nih.gov/). These sequences
were isolated and amplified from total genomic DNA of maize by PCR methods. The
sequences obtained were labeled with biotin and digoxigenin
by PCR as well by enzymatic methods. For the latter we used enzymatic kits: BioNick Labeling System (Invitrogen) to label with biotin
and Dig High Prime (Roche) to label with digoxigenin.
FISH was performed according to Gonz�lez et al. (Chrom.
Res. 14: 629-635, 2006).
FISH slides were observed with a Zeiss AxioPhot epifluorescence microscope (Carl Zeiss, Germany), and
microphotographs were taken with a Leica CCD digital camera.
Chromosomal parameters were measured
using the freeware program MicroMeasure 3.3
(http://www.colostate.edu/depts/ biology/micromeasure).
The relative chromosome length, arm ratio, and centromeric
indexes were calculated to determine the karyotypes. The chromosomes were
ordered from the largest to the smallest, as usual for maize, and chromosome
morphology was described according to Levan et al.
(Hereditas 52: 201-220, 1964). To
estimate the karyotype asymmetry, two numerical parameters were used, according
Romero Zarco (Taxon 35: 526-530, 1986): A1 (intrachromosomal asymmetry index) and A2 (interchromosomal asymmetry index).
����������� In
each race a maximum of 25 individuals were studied (3-5 individuals per maize ear)
and at least 10 cell per individual were analyzed.
The karyotype parameters analysis let
us to elaborate the idiograms from the two maize
races (Figure 1 A and 2A).
DAPI banding allowed
identified and locates the knobs as DAPI-positive bands on mitotic
metaphase chromosomes (Figure 1 B
and 2 B). In FISH experiments, simultaneous hybridization with
the 180-bp and TR-1 probes showed that these sequences co-localized
with all the DAPI-positive bands in both races. Different intensities of
hybridization signals with each probe suggest that the
DAPI-positive knobs are composed by different proportions of 180-bp
and TR-1 sequences.
In table 1 the karyotypic
formulae, A1 and A2 indexes, percentage of heterochromatin and number of knobs are presented for Tupi Amarillo and Rosado maize races.
We observed that the percentage of
heterochromatin of Tupi Amarillo is about 8% higher
than Rosado (Table 1), this difference between both
maize races is due to the higher number of knobs in Tupi Amarillo. This race showed
higher intrachromosomal asymmetry (A1) but lower interchromosomal asymmetry (A2) compared with Rosado. This is due to differences in the size and the
distribution of the knobs on the both
chromosomal arms (Figures 1 A and 2 A).
All results obtained in this work
allowed us to identify cytogenetically the maize races studied. ��������� Then, the variations of the patterns
for number, position and sequence composition of the heterochromatic knobs
are useful markers for a proper cytogenetic characterization of maize races.
The cytogenetic characterization of
different Argentinean races of maize will contribute to the knowledge of the
genetic variability within native materials, useful for its integration in
future breeding plans and biodiversity conservation.
|
|
Tupi�
Amarillo |
�Rosado |
|
Karyotypic formulae |
6m + 4 sm |
6m + 1m-sm + 3sm |
|
% of heterochromatin |
�� 19,64 |
� 11,7 |
|
Range of number of Knobs |
16-22 |
10-11 |
|
A1 |
0,35 |
0,27 |
|
A2 |
0,18 |
0,26 |
Table
1:
Karyotypic parameters of Tupi
Amarillo and Rosado maize races. Ref.: A1: intrachromosomal
asymmetry index. A2: interchromosomal asymmetry
index. m: metacentric. sm: submetacentric.
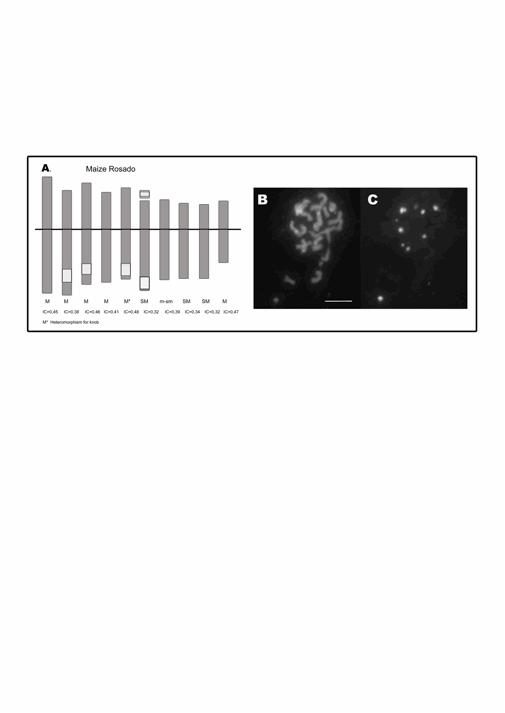
Figure 1: A. Idiogram of maize Rosado. The white blocks
represent the coincident DAPI-positive band and 180-bp and TR-1 FISH signals. B. DAPI-banding.� C.
FISH using 180-pb as probe on mitotic metaphase chromosomes. Ref.: M:
metacentric. SM: submetacentric. IC: centromeric index.� Bar 10 �m.
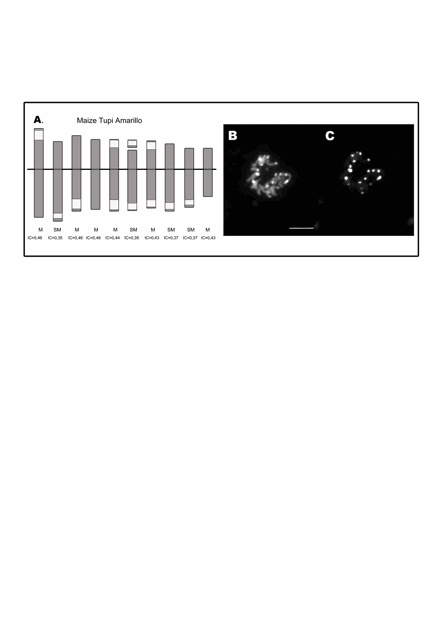 Figure 2: A. Idiogram of maize Tupi Amarillo. The white blocks
represent the coincident DAPI-positive band and 180-bp and TR-1 FISH signals. B. DAPI-banding.� C.
FISH using TR-1 as probe on mitotic metaphase chromosomes. Ref.: M:
metacentric. SM: submetacentric. IC: centromeric index.� Bar 10 �m.
Figure 2: A. Idiogram of maize Tupi Amarillo. The white blocks
represent the coincident DAPI-positive band and 180-bp and TR-1 FISH signals. B. DAPI-banding.� C.
FISH using TR-1 as probe on mitotic metaphase chromosomes. Ref.: M:
metacentric. SM: submetacentric. IC: centromeric index.� Bar 10 �m.
Please
Note: Notes submitted to the Maize Genetics Cooperation Newsletter may be cited
only with consent of authors.