Maize Genetics Cooperation Newsletter vol 85 2011
Johnston, Iowa
Pioneer Hi-Bred, a DuPont
company
Circadian regulation of maize transcriptomes
in B73 and Mo17 inbreds and their reciprocal hybrids.
-- Kevin R. Hayes,
Mary Beatty, Xin Meng, Carl R. Simmons, Jeffrey E. Habben, Olga N. Danilevskaya
Circadian rhythms have been shown
in nearly all forms of life, ranging from algae to plants to mammals. These rhythms include oscillations of
gene expression. We recently surveyed
the maize transcriptome for diurnal rhythmicity and found >23% of all leaf measured transcripts
displaying diurnal regulation under field conditions at the R1 stage of
development [1]. This diurnal
study involved environmental light and temperature variation and thus was different
from classical �circadian� studies, which typically use constant conditions of light
and temperature.
A molecular clock model was recently proposed that links heterosis to the improved or better-tuned circadian clock in
Arabidopsis hybrids [2]. To assess
this hypothesis in maize and to examine classical circadian transcription, leaf
tissues of inbred lines B73 and Mo17 along with their reciprocal hybrids (BxM and MxB) were assayed under constant
conditions over three days by microarray mRNA profiling.
Briefly, plants were grown to the V3 stage
under normal greenhouse conditions and then transferred to a growth chamber
with constant light and temperature (25�C). After 24 hours of acclimation, leaves from three plants per
genotype were collected every four hours for 72 hours. Additionally, B73 whole-root
tissue was collected at each time point.
Messenger RNA was prepared from each genotype~time
point and assayed using an Agilent microarray representing ~43.000 ESTs. The data were normalized via the Quantile
Normalization method and assessed for rhythmicity via
the GeneTS package in the R Statistical Programming
Language as previously described [1].
The leaf results indicate that overall the
number of detected circadian cycling transcripts was lower than in leaves from the
field diurnal study [1], which may be due to the absence in the controlled
environment of other environmental cues that reinforce the diurnal pattern (Table
1). Mo17 leaf tissue displayed
slightly lower numbers of cycling genes and lower amplitude waves than B73,
which may in part be due to probe affinity as the oligos
on the microarray were designed largely according to the B73 genome. There is evidence of a slight attenuation
or �run down� of the cycling pattern, because the median peak/trough values
decreased each successive day. However the third day median peak/trough values
were still approximately two-fold different, indicating good persistence of
circadian rhythms (Table 1 and all Figures). No significant numbers of cycling transcripts were detected
in roots.
Both reciprocal hybrids display markedly
lower numbers of cycling transcripts than the parents (Table 1). The
possibility exists for allelic offsets in peak times to cause broader or lower
amplitude waves that by additive interference diminish those scored as cycling
in hybrids. To resolve this issue shorter time period resolution and
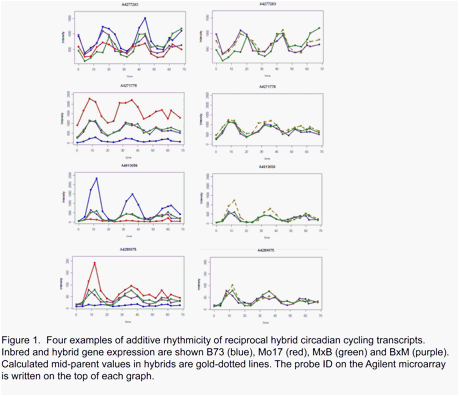
allelic specific expression should be used. Among those cycling transcripts that do appear to cycle in hybrids, they tend
to show an additive pattern between both inbreds
(Figure 1). In many cases the hybrid diurnal patterns could be mimicked
by taking the arithmetic means of the signal from the parent inbreds (Figure 1). In other words the amplitudes of
the hybrid cycling genes correspond to the mid-parent values. The
mid-parent values of cycling transcripts, and the reduced overall number of
cycling transcripts, both indicate an additive pattern likely prevails in
hybrids.
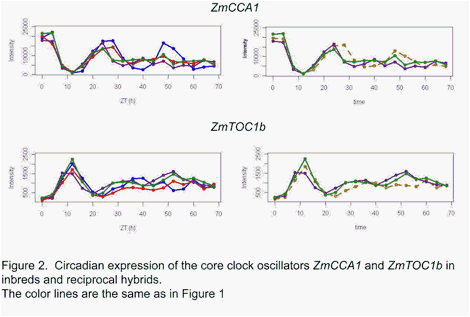
Maize core clock oscillators such as ZmCCA1 and ZmTOC1b (Figure 2) did not show consistent increased amplitude
patterns in the reciprocal hybrids as predicted by the molecular clock model of
heterosis for Arabidopsis [3]. Rather, the circadian clock oscillators
also appeared to more closely match the mid-parent value in hybrids as observed
for the other circadian cycling transcripts.
These results do not support the model for
the circadian machinery having a major impact on heterosis
in maize. The results also
indicate that rhythmic gene expression patterns appear to be mostly additive and do not show epistatic
interactions. These results are consistent with other studies that showed
additive gene expression as the major pattern observed in RNA profiling
experiments of maize hybrids [4, 5].
1. Hayes KR, Beatty M, Meng X, Simmons CR, Habben JE,
et al. (2010) Maize global transcriptomics
reveals pervasive leaf diurnal rhythms but rhythms in developing ears are largely
limited to the core oscillator. PLoS ONE 5(9):
e12887. doi:10.1371/journal.pone.0012887
2. (2009) Altered circadian rhythms regulate growth vigour
in hybrids and allopolyploids. Nature 457: 327–331.
3. Chen ZJ. (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends
Plant Sci. Feb;15(2):57-71.
4. Guo M, Rupe MA, Yang X, Crasta O, Zinselmeier C, Smith OS, Bowen B. (2006) Genome-wide
transcript analysis of maize hybrids: allelic additive gene expression and
yield heterosis. Theor Appl Genet. 113 (5):831-45.
5. Springer NM, Stupar RM. (2007) Allelic
variation and heterosis in maize: how do two halves
make more than a whole? Genome Res.17(3):264-75.
Table 1.
Number of diurnally cycling genes and their median
amplitude at 30% FDR (false
discovery rate)
|
Genotypes |
Number of Cycling Genes |
Median Amplitude (Fold) |
|
B73 |
2191 |
2.44 |
|
Mo17 |
1549 |
1.74 |
|
BxM |
465 |
1.92 |
|
MxB |
203 |
1.97 |
Please Note: Notes submitted to the Maize
Genetics Cooperation Newsletter may be cited only with consent of authors.