Maize Genetics Cooperation
Newsletter vol 85 2011
An endosperm enzyme catalyzes the formation of phosphotriester and phosphodiester bonding complex between nucleic acids with altering their structures
David Pan
University of Wisconsin
Madison
A newly discovered enzyme, that can catalyze the alteration of the structure of nucleic acid through the formation of phosphotriester and phosphodiester bonding complex, was partially purified from maize developing endosperms by combining following sequential
steps: 15% - 35% ammonium sulfate fractionation, DEAD-cellulose anion exchange column chromatography and Sephadex G 150 gel filtration. Endosperms of W64A maize after 22 days of pollination was used for this study. Routinely 50 mM of Tris-HCL buffer, pH 7.5 was used for preparation of enzyme extract, ammonium sulfate fractionation, DEAD anion exchange column chromatography and Sephadex G 150 gel filtration, except that on DEAE-cellulose anion exchange column chromatography, the enzyme was stepwise eluted out from the column by 50 mM, 100 mM, 200 mM of sodium chloride included in Tris buffer, respectively. The enzyme activities were found in both 50 mM and 100 mM eluents. The enzyme activity was monitored by the changing of OD at 260 nm as a result of the formation of a complex of either phosphotriester and phosphodiester bonding between nucleic acids (Fig.1).
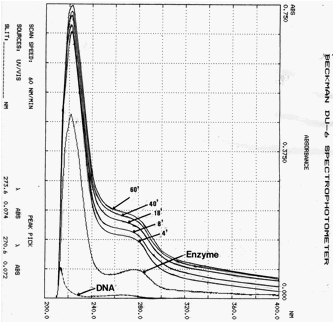
Fig.1. Changing spectra of
the reaction products as a function of time. DNA(only), Enzyme(only)
The optimum pH value for the enzyme activity is in 50 mM acetate buffer at pH 5.4. The enzyme is widely distributed in nature ranging from biological tissues to viral particles including barley mosaic virus, southern mean mosaic virus as well as poliovirus. The broad presence of this enzyme in biological kingdom suggests that the enzyme is an evolution significant protein. A variety of short chain length nucleotides and poly nucleotides including polyU, polyC and polyA had been tested as substrates for demonstrating the catalysis of enzyme reaction. However, the minimum requirement of at least 3 to 4 unites of nucleotides chain length in order to be able initiating the enzyme action suggests that the only phosphate group linked between two nucleotides could not be used to link with other phosphate group from other nucleotide through ester bonding (Figures 2 and 3).
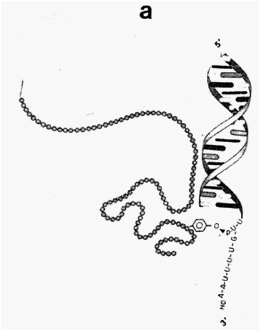
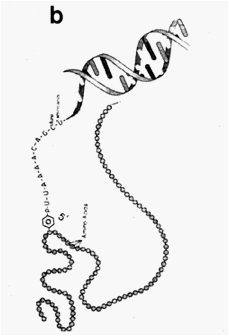
Fig. 2 (a). Enzyme-nucleic acid complex via (b) Enzyme-nucleic acid complex via
phosphotriester bonding phosphodiester bonding
It was found that the enzyme can exist in both monomer (24,000 KD) and dimer (50,000 KD) forms on Sephadex G 150 gel filtration. Both enzyme forms can catalyze the reaction. However, it could not rule out that the enzyme can re-associate into dimer from the monomer form in the reaction mixture before catalysis. The data of 1% agarose gel electrophoresis in 50 mM Tris-HCL buffer at pH 7.5 of the enzyme reaction products from either with long or short chain length of nucleotides as substrate consistently showed a very larger molecular form staying at the origin loading well without migration (data not shown). This together with the preliminary results of electron microscopy indicate that the enzyme can carry out the linking of multi-nucleotides together through either phosphotriester and/or phosphodiester bonding and the formation of a very larger molecular structure (aggregated form). Preliminary study also showed the presence of this high molecular structure of nucleic acid complex, although the detail of structures is not known at the present time, in maize endosperms and Arabiadopsis leaves. A simple overall enzyme reaction mechanism can be depicted as follows:
Nucleotides + Enzyme <-------------> Enzyme-Nucleotide(s) complex (Figure 2)
nEnzyme-Nucleotide(s) <-- -------> nEnzyme + ﴾ Nucleotide(s)-Nucleotide(s)﴿n
Complex through phosphotriester & -diester bondings
This investigation was carried out in the late Dr. Oliver E. Nelson�s laboratory, Department of Genetics, on and off almost two decades with his continuing support and encouragement is a life time memorable relationship between him and the author. Understanding the detail molecular structure of the enzyme reaction products with different chain length of nucleotides as substrates is under investigation.
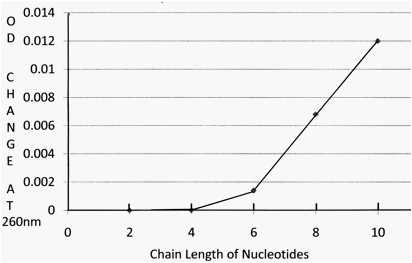
Fig. 3. Minimum requirement of nucleotide chain length (number of nucleotide
Units)
Please Note: Notes submitted to the Maize Genetics Cooperation
Newsletter may be cited only with consent of authors.