Maize Genetics
Cooperation Newsletter vol 85 2011
BERGAMO, Italy1
Unit� di Ricerca per la Maiscoltura – CRA
PADOVA,
Italy2
Dipartimento
di Biotecnologie, Facolt� di Agraria – Universit� di Padova
VERONA, Italy3
Functional
Genomics Center, Dipartimento di Biotecnologie – Universit� di Verona
Identification of cold stress and sulfate starvation
induced microRNAs in maize roots
-
- Altana, A1; Mainieri, D1; Stevanato, P2; Tononi,
P3; Michelotti V1; Ferrarini, A3; Cacco G2;
Rossi, V1
The maize root system plays an essential role in
mediating plant interaction with environmental stimuli. It has been shown that
epigenetic mechanisms and small RNAs (sRNAs) are involved in mediating
transcriptome changes induced by various environmental stresses. In particular,
microRNAs (miRNAs) act in negatively regulating the mRNA level of target genes
(Sunkar
et al, Trends in Plant Science 12: 301-308, 2007; Zhang et al. PLoS Genetics 5:
e716, 2009). To analyze the contribution of miRNAs regulation of
gene transcription in maize roots and in response to abiotic stresses we used
the microarray platforms previously described (Altana et al. a, MGC Newsletter,
this issue). In particular, we analyzed changes in the miRNA root transcriptome
induced by sulfate starvation and cold stress.
For the investigation regarding the sulfate starvation
we employed two maize inbred lines: Lo964 and Lo1016, which are known to differ for root
traits (Sanguineti et al., Maydica 43: 211-216, 1998) with the aim to analyze
the genotype influence on the sulfate starvation mediated modification of miRNAs
population. The protocol for sulfate starvation was established in
hydroponically grown plants, by concomitantly monitoring sulfate uptake rate
(in roots and in leaf) and root architectural parameters. The selected protocol
(13 d-old plants grown 48 h without sulfate: hereinafter named: �-S� and with 500
�M sulfate: hereinafter named �+S) allowed to achieve sulfate starvation
without significantly altering the growth rate of +S vs –S plants. The
RNA was extracted by the primary root because the primary root diameter was one
of the traits that significantly differ between Lo964 and Lo1016 lines. Three
biological replicates were used for extraction of sRNA enriched fraction from
each sample and Cy5 labeled RNA were employed for Combimatrix CustomArray 4X2K
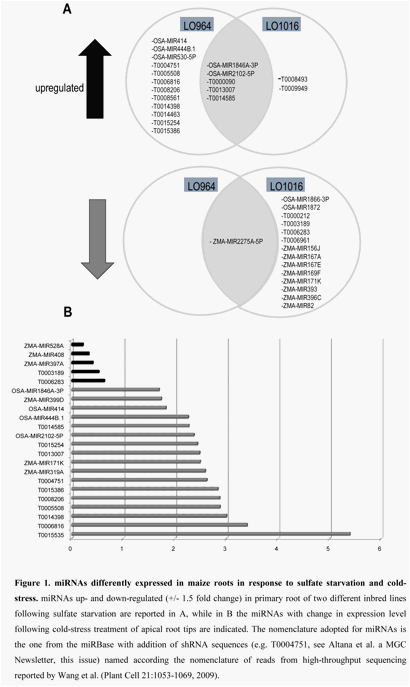
hybridization. Results showed that 18 and 22 miRNAs were differentially
expressed in Lo964-S vs Lo964+S and in Lo1016-S vs Lo1016+S, respectively (a 1.5
fold change, between match probes in different sample types and between
match/mismatch probes within the same sample type, was considered). Six and 18
miRNAs exhibited different abundance when the comparison was between Lo964+S vs
Lo1016+S and Lo964-S vs Lo1016-S. Six miRNAs were commonly affected (5 up- and
1 down-regulated) by sulfate starvation in both lines (Figure 1A). These
observations indicate that the major number of miRNAs changes in response to
sulfate starvation than to genotype. However, because 17 of the 18 sulfate
induced miRNAs are up-regulated in Lo964, whereas most of the miRNAs were
down-regulated in Lo1016 (Figure 1A), a possible genotype-dependent miRNA transcriptome response to sulfate starvation may
occurs. The 6 miRNAs differently expressed in both lines may be instead
part of a genotype-independent conserved sRNA-related mechanism, activated by
sulfate starvation to modulate gene expression.
To investigate miRNA response to cold stress, hydroponically
grown plants were subjected to a cycle of six cold-pulses (13-d old plants were
submitted to 6 days of growth at 25�C for 14 h followed by 10 h where the
temperature dropped to 4 �C). Apical root tips were collected after that both
cold stressed (CS) and not stressed (NS) samples were grown at 25�C for one
day. Three biological replicates were used for each sample and the same
approach described for sulfate starvation was used for miRNA microarray
hybridization and data analysis. The results are illustrated in Figure 1B (22
miRNAs differentially expressed in CS vs NS). The mRNA transcriptome was also
assessed for CS vs NS samples (false discovery rate < 0.05; only differences
with a fold change > +/- 2 were considered). Because we know the predicted target genes of the
differentially expressed miRNAs (Altana et al. a, MGC Newsletter,
this issue) and because miRNAs are expected to negatively regulated expression of
their targets, we searched for mRNA probes exhibiting a negative correlation
with changes in the level of cold-induced miRNA expression. Seventeen miRNAs
up-regulated and 5 miRNAs down-regulated by cold stress negatively correlated with
the signal of 30 and 7 mRNA probes, respectively. The analysis of Gene ontology
(GO) terms for genes represented by these mRNA probes indicated that specific
GO categories were enriched within predicted miRNA targets (phosphate
transmembrane transport, glycerol metabolism, and microtubule-based movement),
thus providing information about possible specific functions for the miRNA-mediated gene regulation in response to
cold stress.
Figure Legend
Figure 1. miRNAs
differently expressed in maize roots in response to sulfate starvation and
cold-stress. miRNAs up- and down-regulated (+/- 1.5 fold change) in primary root of
two different inbred lines grown under sulfate starvation are reported in A,
while in B the miRNAs with change in expression level after cold-stress
treatment of apical root tips are indicated. The nomenclature adopted for
miRNAs is the one from the miRBase, with addition of shRNA sequences (e.g.
T0004751, see Altana et al. a MGC Newsletter, this issue) that are named
according the nomenclature of reads from high-throughput sequencing reported by
Wang et al. (Plant Cell 21:1053-1069, 2009).
Please Note: Notes submitted to the Maize Genetics
Cooperation Newsletter may be cited only
with consent of authors.