Maize Genetics Cooperation
Newsletter vol 84 2010
Please Note: Notes
submitted to the Maize Genetics Cooperation Newsletter may be cited only with
consent of authors.
A small gene family in maize encodes a family of b-glucosidase
aggregating factor (BGAF)-like proteins, and the product of the bgaf2
gene also aggregates b-glucosidase
Farooqahmed S. Kittur1,
David R. Bevan2 and Asim Esen1*
From Department of Biological
Sciences and Biochemistry
Virginia Polytechnic Institute and State University,
Blacksburg, VA 24061-0406
Corresponding author: Asim Esen,
Department of Biological Sciences
Virginia Polytechnic Institute and State University,
Blacksburg, VA 24061-0406
E-mail: [email protected]
In certain maize genotypes called �null�, b-glucosidase,
a major defense related enzyme, forms large insoluble aggregates when tissue
integrity is compromised (Biochem. Genet. 28:31-36, 1990). We have shown that a protein called b-glucosidase
aggregating factor (BGAF) is responsible for b-glucosidase aggregation
and hence the b-glucosidase
null-phenotype (Plant Physiol. 122:563-572, 2000). BGAF is a modular protein
containing an N-terminal dirigent domain and a C-terminal jacalin-related
lectin (JRL) domain (J. Biol. Chem. 282:7299-7311, 2007). BGAF is a lectin; it
shows high preference for galactose but binds other carbohydrates as well. BGAF specifically interacts with maize b-glucosidases
(isozymes Glu1 and Glu2), forming large insoluble complexes (J. Biol. Chem.
282:7299-7311, 2007). Aggregation of b-glucosidase by BGAF does
not affect enzyme activity (unpublished results) nor does bound b-glucosidase
interfere with the ability of BGAF to bind carbohydrates.
Proteins
sharing sequence similarity and modular architecture with BGAF are also
reported from wheat, rice, barley, sorghum and creeping bentgrass Agrostis stolonifera. In wheat, rice, and barley, a small
family of genes encodes BGAF-like proteins, four genes each in wheat and rice,
and three in barley. The products of these genes from wheat (Plant Physiol.
Biochem. 43:185-192, 2005; Plant Physiol. 147:1412-1426, 2008) and one of the
genes from rice (Toxicon. 47:133-139, 2006) are shown to be lectins with
monosaccharide preference for mannose. No protein-aggregating activity was,
however, reported for these proteins. Recently, we showed that an ortholog of
BGAF from sorghum is a GalNAc-specific lectin, but it lacks protein-aggregating
activity (Glycobiol. 19:277-287, 2009). Although the above observations suggest
that the b-glucosidase
aggregating activity is unique to maize BGAF alone, the occurrence of other
BGAF-like proteins in maize with protein aggregating activity cannot be ruled
out. We predicted that multiple genes encoding BGAF-like proteins must be
present in maize also, since its close relatives wheat, rice, and barley each
have a small family of genes encoding BGAF-like proteins.
Searching
the maize genome database (www.maizeGDB)
using the maize BGAF cDNA sequence as query led to identification of at least
six genes. The gene encoding BGAF, which we described earlier (J. Biol. Chem.
282:7299-7311, 2007) is located on chromosome 7 and was designated as bgaf1. The remaining genes were denoted
as bgaf2, bgaf3, bgaf4, bgaf5 and bgaf6, where the order reflects their divergence distance in
sequence from bgaf1. One gene (bgaf 7) predicted from EST sequences was
not found in the maize genome sequence. The remaining genes are located on
chromosomes 2 (bgaf2), 6 (bgaf3, bgaf4, and bgaf5) and 8 (bgaf6), respectively.
The predicted protein products of genes bgaf2, bgaf3, bgaf4, bgaf5 and bgaf6 share 69%, 44%, 45%, 43%, and 41% sequence identity, respectively,
with BGAF1, and vary in size from 32 to 35 kD. Maize EST
database searches identified EST clones AY105022, AY104689, AY103569, BT016225
and BT042436 whose sequences matched with the sequences of bagf2, bgaf3,
bgaf4, bgaf5, and bgaf6 genes, respectively. Moreover, we were able to construct
complete cDNA coding sequences corresponding to seven different bgaf genes including bgaf7 using sequence overlaps among EST
clones in the database. To investigate if the products of these genes (other
than bgaf1) participate in
protein-protein interactions, an EST clone (AY105022) corresponding to the bgaf2 gene was selected. Complete
sequencing of EST AY105022 indicated that it contained a 948 bp open-reading
frame encoding a 315 amino acid long polypeptide, consisting of a predicted
N-terminal dirigent domain and a C-terminal JRL domain. The predicted protein
was identical to the product of bgaf2
gene located on chromosome 2.
Both full-length BGAF2 protein and its JRL domain (produced
in E. coli) showed binding to maize
Glu1. In the pull-down assay, precipitable complexes of maize Glu1 were
obtained with full-length protein, whereas no such complexes were observed with
its JRL domain. In the gel-shift assay, in the presence of full-length protein,
the Glu1 activity zone showed smearing, extending from the sample well to the
boundary between stacking and resolving gel (Fig. 1, lane 4), indicating
formation of large aggregates. The JRL domain also showed a distinct activity
band of mobility slower than Glu1, suggesting formation of a smaller, soluble
JRL-Glu1 complex (Fig 1, lane 5). The above results clearly indicate that the
product of the bgaf2 gene is also a b-glucosidase aggregating factor, which we designated as
BGAF2.
To
investigate whether BGAF2 is expressed in maize, null-line H95 shoots were
extracted with PBS (pH 7.4) and the BGAF2-b-glucosidase complexes were isolated after passing through
a column of Nickel with immobilized rGlu1 on it, followed by affinity
chromatography on lactosyl-agarose. SDS-PAGE of fractions obtained from the
lactosyl-agarose column showed the presence of four bands of sizes 62, 60, 34
and 32 kD, respectively (Fig. 2, lane 4). The 60 and 62 kD bands are native and
recombinant maize Glu1, respectively. The latter (rGlu1) is larger than native
Glu1 because of the presence of a His-tag. The 32 kD band is native BGAF1.
The ~34 kD protein band was
found to be immuno-reactive with antisera raised against a BGAF2 specific
peptide, suggesting that the 34 kD protein is in fact a BGAF-like protein,
namely BGAF2. To establish the identity of the 34 kD protein band
unequivocally, it was excised from the gel and subjected to LC-MS/MS analysis.
Three peptides of sequence ANQAAILESK, FSGSTLEVR and
VGPWGGSGGPMELTETETPMR were identified from LC-MS/MS
analysis. When these were used as query to search the current (Nov 2008)
release of the NR (NCBI) database, the product of the
bgaf2 gene received the highest hits,
indicating that the 34 kD protein band is in fact BGAF2.
The b-glucosidase null phenotype in maize (H95 null-line)
is not due to BGAF1 alone. BGAF2 is also responsible for b-glucosidase aggregation although its contribution is
minor compared to BGAF1 because it is less abundant than BGAF1. There are at
least eleven maize null-lines in which we have observed that BGAF1 is a major
factor responsible for b-glucosidase aggregation
(unpublished data). It is not known at this time whether bgaf3, bgaf4, bgaf5 and bagf6
genes are expressed in maize at the protein level, and if so, whether their
products participate in protein-protein interaction. The fact there are cDNAs
corresponding to each of the seven predicted bgaf genes in the maize EST database indicates that these genes are
transcribed and show both temporal and spatial expression patterns. For
example, bgaf1 and bgaf2 are expressed in aerial organs,
whereas bgaf4, bgaf5, and bgaf6 are expressed in embryo and
endosperm, respectively (http://www.ncbi.nlm.nih.gov/unigene).
The precise physiological role of b-glucosidase-BGAF
interactions is not well understood at this time. We speculate that these
interactions have a key function in plant defense responses. There is good
reason to believe that these lectins deter the insect larvae from feeding onto
plants by lodging the enzymes in the oral cavity or the midgut (by binding to
glycoproteins) and causing a localized burst of toxic chemicals (aglycones and
their break-down products) in these cavities. In fact, the protein product of
the Hfr-1 gene, a BGAF homolog from
wheat, has been shown to deter Hessian fly larvae from feeding on resistant
plants by binding to sensory receptors (Plant Physiol. 147:1412-1426, 2008). It
is now becoming clear that there are at least two chimeric lectins in maize
that specifically interacts with β-glucosidases. We postulate they help
the plant to launch a powerful defense response to attack by pests.
1 2 3 4 5
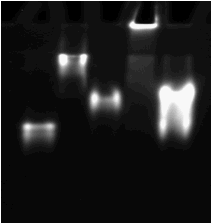
Fig. 1. Gel-shift assay to detect binding
of recombinant BGAF2 and its JRL domain to maize Glu1. Maize Glu1 (125 nM)
was mixed with BGAF1 (150 nM), its JRL domain (500 nM), BGAF2 (400 nM), and its
JRL domain (10 mM),
and incubated at room temperature for 30 min. Following incubation, 40 ml of
reaction mixtures were mixed separately with 20 ml of sample buffer and electrophoresed
on a 8% native gel. b-Glucosidase
activity was detected by incubating gel with 4-methylumbelliferyl-b-D-glucopyranoside.
Lane 1, Glu1; lane 2, Glu1+BGAF1; lane 3,
Glu1+JRL domain of BGAF1; lane 4,
Glu1+BGAF2; lane 5, Glu1+the JRL
domain of BGAF2.
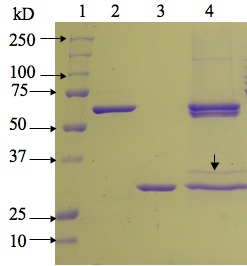
Fig. 2. SDS-PAGE profile of complexes of
maize b-glucosidase with BGAF1
and BGAF-like protein isolated from maize null-line H95. Lane 1, Molecular weight markers; lane 2, recombinant maize Glu1 expressed
with His-tag; lane 3, recombinant
BGAF1; lane 4, b-glucosidase-BGAF
complexes isolated from maize null-line H95. Note that there are four bands in
lane 4, two of which are native (60 kD, lower band) and recombinant (62 kD,
upper band) Glu1, respectively. The protein band of size similar to rBGAF1 is
native BGAF1. The band (34 kD) immediately above native BGAF1,
which was immuno-reactive with BGAF2 specific antibody, was excised and
subjected to LC-MS/MS analysis to establish its identity.